1 TL;DR
Out in the real world you may often find yourself working with data from multiple sources. It will probably be stored in separate files and you’ll need to combine them before you can attempt to answer any of your research questions. This post will show you how you can combine data frames using another set of dplyr functions called joins.
2 Introduction
The 6th post of the Scientist’s Guide to R series is all about using joins to combine data. While tidy data organized nicely into a single .csv or .xlsx spreadsheet may be provided to you in courses, in the real world you’ll often collect data from multiple sources often only containing one or two similar “key” columns (like subject ID #) and have to combine pieces of them to do anything interesting. This type of data is called relational data, since the datasets are related through common key column(s). Relational databases are how most data are stored in modern non-academic organizations.
Fortunately, a package we’re already familiar with from a couple of posts ago, dplyr, also has a set of functions for combining data with functions called “joins”. For this post we will cover the 6 most common joins:
left_join(x, y)which combines all columns in data framexwith those in data frameybut only retains rows fromx.right_join(x, y)also keeps all columns but operates in the opposite direction, returning only rows fromy.full_join(x, y)combines all columns of x with all columns of y and retains all rows from both data frames.inner_join(x, y)combines all columns present in eitherxorybut only retains rows that are present in both data frames.semi_join(x, y)returns the columns fromxonly and retains rows ofxthat are also present inyanti_join(x, y)returns the columns fromxonly and retains rows ofxthat are not present iny.
A nice design feature of these functions is that their names and behaviour were inspired by analogous functions for joining data in the ubiquitous database management programming language “Stuctured Query Language” (SQL). Learning dplyr therefore also makes SQL easier to learn in the future, which will be helpful if you ever want to work with data for a living.
In case you find yourself working in an environment where only base R is available, we’ll also cover how to join data using the base R function merge().
Aside from specifying the data frames to be joined, one other thing we need to do is specify the key column(s) to be used for aligning the rows prior to joining the data.
Key columns are specified with the by argument, e.g. inner_join(x, y, by = "subject_id") adds columns of y to x for all rows where the values of the “subject_id” column (present in each data frame) match. If the name of the key column differs between the data frames, e.g. “subject_id” in x and “subj_id” in y, then you have to specify both names using by = c("subject_id" = "subj_id") so that the functions know which columns to compare.
A nice feature of the *_join() functions is that if you don’t specify the by argument they will assume that columns with the same names across x and y are key columns. This is very convenient when the columns with the same names in fact contain the same type of values.
2.1 setup
To demonstrate the use of the join functions, I’ll prepare an example of relational data using the gapminder dataset for the year 2007 aggregated to the continent level. In this representation of the data, the life expectancy, population, and gdpPercap are stored in separate data frames (called life_df, pop_df, and gdp_df respectively). This sort of arrangement is closer to what you might encouter if the gapminder data were stored in a relational database.
library(gapminder) #contains the gapminder data
library(dplyr) #functions for manipulating and joining data
life_df <- gapminder %>%
filter(year >= 1997 & year <= 2007) %>%
group_by(continent, year) %>%
summarise(mean_life_expectancy = mean(lifeExp, na.rm = T)) %>%
filter(continent != "Asia") %>% ungroup()
pop_df <- gapminder %>%
filter(year >= 1997 & year <= 2007) %>%
group_by(continent, year) %>%
summarise(total_population = sum(pop, na.rm = T)) %>%
filter(continent != "Europe") %>%
ungroup()
gdp_df <- gapminder %>%
filter(year == 1997 | year == 2007) %>%
group_by(continent, year) %>%
summarise(total_gdpPercap = sum(gdpPercap, na.rm = T)) %>%
ungroup()Recall that we can print a view of the structure of each data frame using the glimpse function from dplyr
life_df %>% glimpse()## Rows: 12
## Columns: 3
## $ continent <fct> Africa, Africa, Africa, Americas, Americas, Ameri~
## $ year <int> 1997, 2002, 2007, 1997, 2002, 2007, 1997, 2002, 2~
## $ mean_life_expectancy <dbl> 53.59827, 53.32523, 54.80604, 71.15048, 72.42204,~pop_df %>% glimpse()## Rows: 12
## Columns: 3
## $ continent <fct> Africa, Africa, Africa, Americas, Americas, Americas,~
## $ year <int> 1997, 2002, 2007, 1997, 2002, 2007, 1997, 2002, 2007,~
## $ total_population <dbl> 743832984, 833723916, 929539692, 796900410, 849772762~gdp_df %>% glimpse()## Rows: 10
## Columns: 3
## $ continent <fct> Africa, Africa, Americas, Americas, Asia, Asia, Europe~
## $ year <int> 1997, 2007, 1997, 2007, 1997, 2007, 1997, 2007, 1997, ~
## $ total_gdpPercap <dbl> 123695.50, 160629.70, 222232.52, 275075.79, 324525.08,~or print them to the console using the names
life_df## # A tibble: 12 x 3
## continent year mean_life_expectancy
## <fct> <int> <dbl>
## 1 Africa 1997 53.6
## 2 Africa 2002 53.3
## 3 Africa 2007 54.8
## 4 Americas 1997 71.2
## 5 Americas 2002 72.4
## 6 Americas 2007 73.6
## 7 Europe 1997 75.5
## 8 Europe 2002 76.7
## 9 Europe 2007 77.6
## 10 Oceania 1997 78.2
## 11 Oceania 2002 79.7
## 12 Oceania 2007 80.7pop_df ## # A tibble: 12 x 3
## continent year total_population
## <fct> <int> <dbl>
## 1 Africa 1997 743832984
## 2 Africa 2002 833723916
## 3 Africa 2007 929539692
## 4 Americas 1997 796900410
## 5 Americas 2002 849772762
## 6 Americas 2007 898871184
## 7 Asia 1997 3383285500
## 8 Asia 2002 3601802203
## 9 Asia 2007 3811953827
## 10 Oceania 1997 22241430
## 11 Oceania 2002 23454829
## 12 Oceania 2007 24549947gdp_df ## # A tibble: 10 x 3
## continent year total_gdpPercap
## <fct> <int> <dbl>
## 1 Africa 1997 123695.
## 2 Africa 2007 160630.
## 3 Americas 1997 222233.
## 4 Americas 2007 275076.
## 5 Asia 1997 324525.
## 6 Asia 2007 411610.
## 7 Europe 1997 572303.
## 8 Europe 2007 751634.
## 9 Oceania 1997 48048.
## 10 Oceania 2007 59620.From these printouts we can tell that each data frame only has values for some of the continents and/or some of the years that are present in the full gapminder data. I’ve structured them this way so that it is easier to see how they are joined, as you’ll soon find out.
3 left_join()
If we wanted to add the population data for each continent that appears in the life expectancy data frame, we could use left_join() and the key columns continent and year.
#all columns in x will be returned and
#all columns of y will be returned
#for rows in the key column that have values in y that match those in x
left_join(x = life_df,
y = pop_df,
by = c("continent", "year"))## # A tibble: 12 x 4
## continent year mean_life_expectancy total_population
## <fct> <int> <dbl> <dbl>
## 1 Africa 1997 53.6 743832984
## 2 Africa 2002 53.3 833723916
## 3 Africa 2007 54.8 929539692
## 4 Americas 1997 71.2 796900410
## 5 Americas 2002 72.4 849772762
## 6 Americas 2007 73.6 898871184
## 7 Europe 1997 75.5 NA
## 8 Europe 2002 76.7 NA
## 9 Europe 2007 77.6 NA
## 10 Oceania 1997 78.2 22241430
## 11 Oceania 2002 79.7 23454829
## 12 Oceania 2007 80.7 24549947# if the key columns have different names, you can tell the join function which
# columns to use with the equality operator
#1st I'll just rename the continent column for pedagogical purposes
life_df_renamed <- rename(life_df,
cont = continent)
names(life_df_renamed)## [1] "cont" "year" "mean_life_expectancy"left_join(x = life_df_renamed,
y = pop_df,
#since the continent column is now called "cont" in life_df,
#we have to tell left_join which columns to match on.
#You'll get an error if you try by = c("continent", "year") this time
by = c("cont" = "continent",
"year"))## # A tibble: 12 x 4
## cont year mean_life_expectancy total_population
## <fct> <int> <dbl> <dbl>
## 1 Africa 1997 53.6 743832984
## 2 Africa 2002 53.3 833723916
## 3 Africa 2007 54.8 929539692
## 4 Americas 1997 71.2 796900410
## 5 Americas 2002 72.4 849772762
## 6 Americas 2007 73.6 898871184
## 7 Europe 1997 75.5 NA
## 8 Europe 2002 76.7 NA
## 9 Europe 2007 77.6 NA
## 10 Oceania 1997 78.2 22241430
## 11 Oceania 2002 79.7 23454829
## 12 Oceania 2007 80.7 24549947Note that the total_population column from pop_df has been joined to life_df based on matching values in the key columns that appear in both data frames. Since we used a left join and Europe is listed as a continent in life_df, the row for it is returned in the joined data frame. However, because there are no population values for Europe in pop_df, these rows are filled with NAs under the total_population column.
4 right_join()
A right join is basically the same thing as a left_join but in the other direction, where the 1st data frame (x) is joined to the 2nd one (y), so if we wanted to add life expectancy and GDP per capita data we could either use:
a
right_join()withlife_dfon the left side andgdp_dfon the right side, ora
left_join()withgdp_dfon the left side andlife_dfon the right side
… and get the same result with only the columns arranged differently…
#Also, since the key columns have the same names in each data frame we don't have to specify them
#we can also pipe in the 1st dataframe using the pipe operator (`%>%`)
rj <- life_df %>% right_join(gdp_df)
rj## # A tibble: 10 x 4
## continent year mean_life_expectancy total_gdpPercap
## <fct> <int> <dbl> <dbl>
## 1 Africa 1997 53.6 123695.
## 2 Africa 2007 54.8 160630.
## 3 Americas 1997 71.2 222233.
## 4 Americas 2007 73.6 275076.
## 5 Europe 1997 75.5 572303.
## 6 Europe 2007 77.6 751634.
## 7 Oceania 1997 78.2 48048.
## 8 Oceania 2007 80.7 59620.
## 9 Asia 1997 NA 324525.
## 10 Asia 2007 NA 411610.lj <- gdp_df %>% left_join(life_df)
identical(rj, lj)## [1] FALSEidentical(rj, select(lj, 1, 2, 4, 3))## [1] FALSEThis time there are missing values for Asia’s mean life expectancy because Asia does not appear in the continent column of life_df (but it does appear in gdp_df), and no rows for the year 2002 because 2002 does not appear in the “year” key column of gdp_df.
5 full_join()
After aligning rows by matches in the key column(s), a full join retains all rows that appear in x or y
life_df %>%
full_join(gdp_df) %>%
arrange(year, continent) #sort by year then by continent## # A tibble: 14 x 4
## continent year mean_life_expectancy total_gdpPercap
## <fct> <int> <dbl> <dbl>
## 1 Africa 1997 53.6 123695.
## 2 Americas 1997 71.2 222233.
## 3 Asia 1997 NA 324525.
## 4 Europe 1997 75.5 572303.
## 5 Oceania 1997 78.2 48048.
## 6 Africa 2002 53.3 NA
## 7 Americas 2002 72.4 NA
## 8 Europe 2002 76.7 NA
## 9 Oceania 2002 79.7 NA
## 10 Africa 2007 54.8 160630.
## 11 Americas 2007 73.6 275076.
## 12 Asia 2007 NA 411610.
## 13 Europe 2007 77.6 751634.
## 14 Oceania 2007 80.7 59620.The output now includes rows for the year 2002, which were present in life_df but not in gdp_df. It also includes rows for Asia, which are present in gdp_df but are missing from life_df. As you can see, a full join retains all of the data, filling in missing values where necessary.
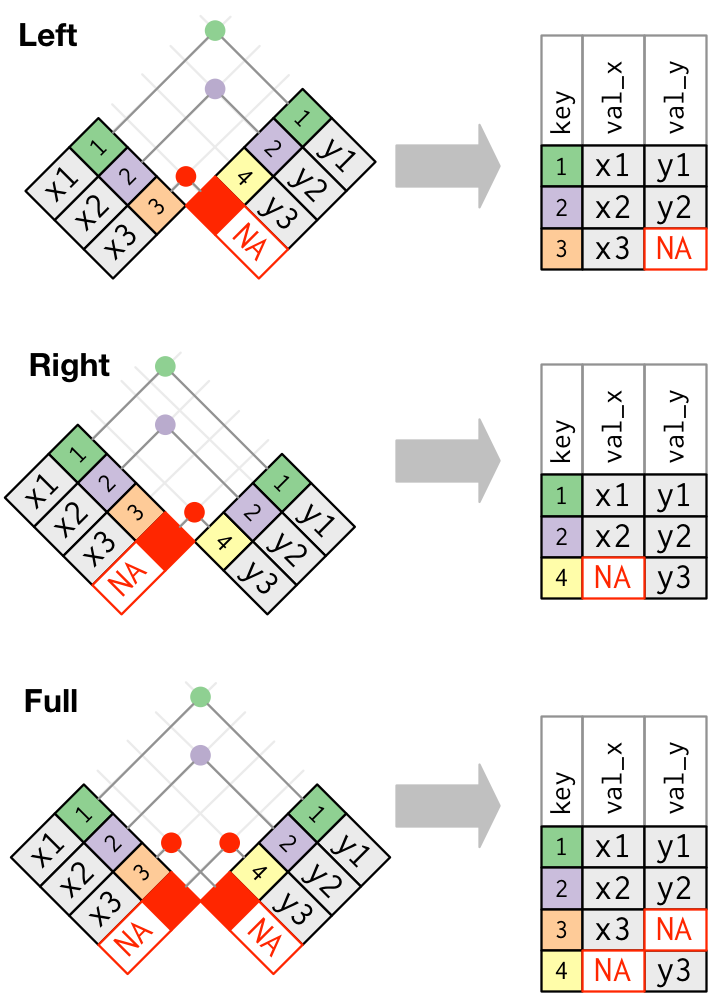
Left joins, right joins, and full joins are also collectively referred to as outer joins because they retain the observations from at least one of the joined tables. This excellent set of diagrams from R for Data Science (R4DS) can help you build an intuitive sense of how these outer joins work:
6 inner_join()
Often you may only want to work with rows which have matching entries in both data sources. Since only some rows are retained, we’re no longer dealing with an outer join. In this case you could use inner_join(), which returns all rows in both x and y but only rows with that appear in the key columns of both data frames.
life_df %>%
inner_join(gdp_df)## # A tibble: 8 x 4
## continent year mean_life_expectancy total_gdpPercap
## <fct> <int> <dbl> <dbl>
## 1 Africa 1997 53.6 123695.
## 2 Africa 2007 54.8 160630.
## 3 Americas 1997 71.2 222233.
## 4 Americas 2007 73.6 275076.
## 5 Europe 1997 75.5 572303.
## 6 Europe 2007 77.6 751634.
## 7 Oceania 1997 78.2 48048.
## 8 Oceania 2007 80.7 59620.This time we only get data for 1997 and 2007 even though life_df has values for 2002 because gdp_df did not contain any data for 2002. We also don’t get any data for Asia, which was present in gdp_df, because there was no data for Asia in life_df.
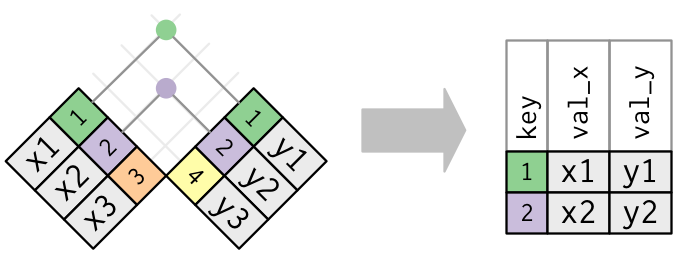
This diagram from R4DS shows you another example of how an inner join works:
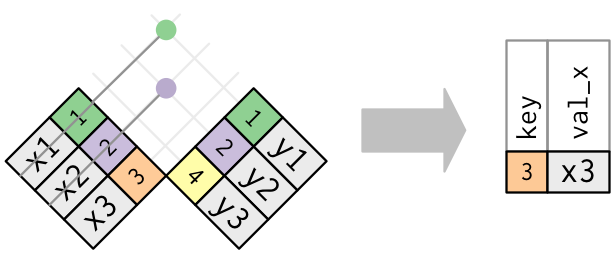
7 semi_join()
So far we’ve been filtering rows based on matches in the key columns but extacting all columns from both data frames. The other two main dplyr join functions are available for situations where you only want to keep the columns of one data frame (x).
semi_join(x, y) filters the rows of x to retain only those that also appear in y
life_df %>%
semi_join(pop_df)## # A tibble: 9 x 3
## continent year mean_life_expectancy
## <fct> <int> <dbl>
## 1 Africa 1997 53.6
## 2 Africa 2002 53.3
## 3 Africa 2007 54.8
## 4 Americas 1997 71.2
## 5 Americas 2002 72.4
## 6 Americas 2007 73.6
## 7 Oceania 1997 78.2
## 8 Oceania 2002 79.7
## 9 Oceania 2007 80.7This time we only get the columns from life_df and we’ve dropped rows for Europe because Europe only appears under the continent key column for life_df and not pop_df.
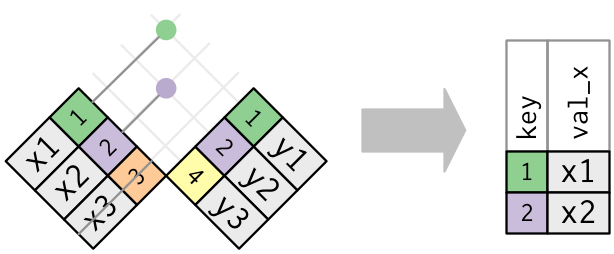
Here is the R4DS semi-join diagram showing that only columns and rows from table 1 are retained as a second example:
8 anti_join()
In contrast to semi joins, anti joins return the rows x that do not appear in y.
life_df %>%
anti_join(pop_df)## # A tibble: 3 x 3
## continent year mean_life_expectancy
## <fct> <int> <dbl>
## 1 Europe 1997 75.5
## 2 Europe 2002 76.7
## 3 Europe 2007 77.6As you can see, anti joins can be very useful if you want to know which rows are excluded due to mismatches in the key columns. Checking for consistencies and inconsistencies between data sources is an important part of the data cleaning process and can often help to uncover data entry or coding errors that should be fixed prior to conducting any analyses.
R4DS diagram showing how the anti-join works:
9 building data frames using bind_rows() or bind_cols()
If you have two data frames with the same columns, you can combine their rows using dplyr::bind_rows() or rbind(). rbind() is best suited for rowwise combinations of vectors or matrices, while bind_rows() is better for combining data frames. In my experience, the most common reason to use either would be to add data for new cases to a data frame, so I will only demonstrate bind_rows() here. For example, say we decided to add the gapminder life expectancy data for Asia to life_df:
#first get the data for asia from the original gapminder dataset
asia_life_exp <- gapminder %>%
filter(continent == "Asia",
between(year, 1997, 2007)) %>% #between is a shortcut for (column >= value & column <= value)
group_by(continent, year) %>%
summarise(mean_life_expectancy = mean(lifeExp, na.rm = T))
#then add it to the top of life_df
bind_rows(asia_life_exp, life_df) ## # A tibble: 15 x 3
## # Groups: continent [5]
## continent year mean_life_expectancy
## <fct> <int> <dbl>
## 1 Asia 1997 68.0
## 2 Asia 2002 69.2
## 3 Asia 2007 70.7
## 4 Africa 1997 53.6
## 5 Africa 2002 53.3
## 6 Africa 2007 54.8
## 7 Americas 1997 71.2
## 8 Americas 2002 72.4
## 9 Americas 2007 73.6
## 10 Europe 1997 75.5
## 11 Europe 2002 76.7
## 12 Europe 2007 77.6
## 13 Oceania 1997 78.2
## 14 Oceania 2002 79.7
## 15 Oceania 2007 80.7If we instead had two data frames with the same cases/rows but different columns (and no common key columns to enable the use of joins), we could combine them using dplyr::bind_cols() or cbind(). Again, bind_cols() is preferred over cbind() for combining data frames by column. However, if you’re only adding a few columns, dplyr::mutate() or a "df$newcol <- newcol" statement per column to add would also work.
For example, if we were provided with each column of the gapminder dataframe as separate vectors (each with values in identical order), without any common key columns among any of the fragments, we could reconstruct the orginal gapminder data frame using bind_cols(), e.g.:
ctry <- gapminder$country
ctin <- gapminder$continent
yr <- gapminder$year
le <- gapminder$lifeExp
pop <- gapminder$pop
gdp <- gapminder$gdpPercap
bound_gap <- bind_cols("country" = ctry, #add names using "name" = vector syntax
"continent" = ctin,
"year" = yr, "lifeExp" = le,
"pop" = pop,
"gdpPercap" = gdp)
identical(gapminder, bound_gap)## [1] TRUE#the main difference between cbind() and bind_rows() is that bind_rows returns a
#tibbleWhen considering the use of rbind()/bind_rows() or cbind()/bind_cols() you must keep in mind that because no key columns are checked for matching values you need to be sure that the columns (when binding rows) or rows (when binding columns) are arranged in exactly the same order for each portion of the dataframe before binding the pieces together. This approach can be very error prone, particularly in cases where data cleaning or analysis is being done collaboratively.
For this reason I strongly recommend that you make use of key columns and combine data using joins whenever possible.
9.1 add_row()
If you only wanted to add a single row to a data frame, you can use tibble::add_row() and (recall that the tibble package is also part of the tidyverse). Let’s say (hypothetically of course) we found out that the mean life expectancy for countries in Africa had gone up to 56 for the year 2012. We could add this row as follows:
updated_life_df <- life_df %>%
tibble::add_row(continent = "Africa", year = 2012, mean_life_expectancy = 56) %>% #specify values to be added using column = value syntax
arrange(continent, year)
updated_life_df #now the value we added appears in the printout of the data frame## # A tibble: 13 x 3
## continent year mean_life_expectancy
## <chr> <dbl> <dbl>
## 1 Africa 1997 53.6
## 2 Africa 2002 53.3
## 3 Africa 2007 54.8
## 4 Africa 2012 56
## 5 Americas 1997 71.2
## 6 Americas 2002 72.4
## 7 Americas 2007 73.6
## 8 Europe 1997 75.5
## 9 Europe 2002 76.7
## 10 Europe 2007 77.6
## 11 Oceania 1997 78.2
## 12 Oceania 2002 79.7
## 13 Oceania 2007 80.710 joining 3 or more data frames
Joining 3 or more data frames is also pretty easy using dplyr, just pipe the output of a join into another join. This is incredibly simple if they all have key columns with the same names. For example, if I wanted to combine life_df, pop_df and gdp_df and keep rows present in any of them, all I have to do is:
combined_data <- life_df %>%
full_join(pop_df) %>% #life_df is inserted as the 1st argument i.e. data frame "x" of the full_join
full_join(gdp_df) #the output of the previous join is passed to the first argument of the second full_join
combined_data #now it's all together, using what could be condensed into a single line of code## # A tibble: 15 x 5
## continent year mean_life_expectancy total_population total_gdpPercap
## <fct> <int> <dbl> <dbl> <dbl>
## 1 Africa 1997 53.6 743832984 123695.
## 2 Africa 2002 53.3 833723916 NA
## 3 Africa 2007 54.8 929539692 160630.
## 4 Americas 1997 71.2 796900410 222233.
## 5 Americas 2002 72.4 849772762 NA
## 6 Americas 2007 73.6 898871184 275076.
## 7 Europe 1997 75.5 NA 572303.
## 8 Europe 2002 76.7 NA NA
## 9 Europe 2007 77.6 NA 751634.
## 10 Oceania 1997 78.2 22241430 48048.
## 11 Oceania 2002 79.7 23454829 NA
## 12 Oceania 2007 80.7 24549947 59620.
## 13 Asia 1997 NA 3383285500 324525.
## 14 Asia 2002 NA 3601802203 NA
## 15 Asia 2007 NA 3811953827 411610.# Alternatively, I could use full_join(life_df, pop_df) %>% full_join(gdp_df)
# or full_join(full_join(life_df, pop_df), gdp_df)This is another example of how nested function calls can be easier to read when written in series with the pipe operator (%>%), which I covered in more detail here.
11 merge()
In the very unlikely event that you find yourself having to combine relational data but are working on a computer that only has base R and no admin priviledges to enable you to install dplyr, have no fear! You can use the merge() function from base R to perform left joins, right joins, inner joins, and full joins.
In addition to the x and y arguments that need to be used to specify the data frames to be joined and the by argument that indicates which key columns to use, the type of join is determined via the all.x and all.y arguments
# inner join/merge
ij_merge <- merge(life_df, pop_df,
all.x = F, all.y = F) #the default merge/join is an inner join, in which all.x and all.y are both FALSE
ij_dplyr <- inner_join(life_df, pop_df) %>%
as.data.frame() #removes the tibble class which the merge result doesn't have
all.equal(ij_merge, ij_dplyr)## [1] TRUE#full join/merge
fj_merge <- merge(life_df, pop_df,
all.x = T, all.y = T) #set (all.x = T and all.y = T) or (all = T), to perform a full join
fj_dplyr <- full_join(life_df, pop_df) %>%
as.data.frame() %>%
arrange(continent)
all.equal(fj_merge, fj_dplyr)## [1] TRUE#for a left join, all.x should be set to TRUE and all.y to FALSE
lj_merge <- merge(life_df, pop_df,
all.x = T, all.y = F)
lj_dplyr <- left_join(life_df, pop_df) %>% as.data.frame()
all.equal(lj_merge, lj_dplyr)## [1] TRUE#for a right join, all.x should be set to FALSE and all.y to TRUE
rj_merge <- merge(life_df, pop_df,
all.x = F, all.y = T)
rj_dplyr <- right_join(life_df, pop_df) %>% as.data.frame()
all.equal(rj_merge, rj_dplyr)## [1] "Component \"continent\": 6 string mismatches"
## [2] "Component \"mean_life_expectancy\": 'is.NA' value mismatch: 3 in current 3 in target"
## [3] "Component \"total_population\": Mean relative difference: 1.974144"Why bother with dplyr joins if merge() can do so much? Simply because the dplyr code is easier to read and the dplyr functions are faster. Unfortunately, merge also can’t handle semi joins or anti joins, so you’d have to do a bit more work to achieve the same results without dplyr.
If you’ve followed along, congratulations! You now know how the basics of combining data frames in with dplyr joins and base R. Just practice these operations a few times on your own and joins will seem trivial!
13 Notes
You can learn more about joins, working with relational data, and the set operation functions in
Rhere.You can learn more about
SQLhere.The author of dplyr, Hadley Wickham, also wrote the dbplyr package, which translates dplyr to SQL for you so you can actually query databases directly using dplyr code or even view the SQL code translations. You can learn more about dbplyr here. The code translation is really helpful if you’re trying to learn SQL.
Thank you for visiting my blog. I welcome any suggestions for future posts, comments or other feedback you might have. Feedback from beginners and science students/trainees (or with them in mind) is especially helpful in the interest of making this guide even better for them.
This blog is something I do as a volunteer in my free time. If you’ve found it helpful and want to give back, coffee donations would be appreciated.